The Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV-2) is a virus in the Corona virus family that causes COVID-19. One fact about viruses is that they have innate ability to mutate and produce variants. In just over a year of the SARS-CoV-2 pandemic, over 20,000 distinct viral mutations have been recorded across the viral genome (1).
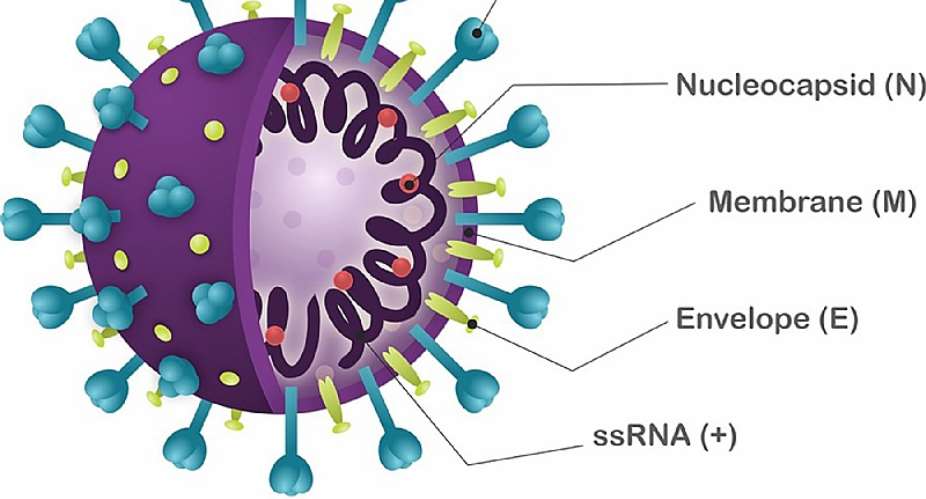
A thorough discussion of the genetics and pathogenesis of the virus was held under the maiden edition of ‘The COVID-19 Diary’ rolled out by the Association of African Universities on August 24, 2021, the panelists explained that when a virus enters a host cell, it makes a copy of itself to infect the cell. For SARS-CoV-2, it is an enveloped spherical virus which has protein that allows it to attach to the host cell. The angiotensin-converting enzyme 2 (ACE2) receptor on the host cell's membrane, serves as a pathway for the genome ribonucleic acid (RNA) to pass through to the host cell and replicate. The RNA combines with a molecular protein in the host cell, the ribosome, to interact the viral RNA, resulting in the production of the virus's replicase and structural proteins (2).
In some cases, errors occur during the replication process, impacting negatively or positively on the virus's ability to survive and replicate. There is the possibility of changes in the virus structure, its ability to affect, its ability to spread, and its ability to cause more severe illness and death. These changes are referred to as mutations, detected through a process known as genome sequencing (3, 4). It is normal for viruses to change and evolve as they spread among people over time, allowing the virus to adapt to its surroundings and move from host to host more effectively.
When a virus' change become significantly different from the original virus, they are referred to as variants. Variants are classified according to specific markers associated with changes in receptor binding; decreased antibody neutralization developed against previous infection or vaccination; and decreased efficacy of treatments. Those with potential diagnostic impact are known as variants of interest (VOI). The second class is known as the variants of concern (VOC). A VOI becomes VOC if it is known to spread more easily; cause more disease severity, including hospitalizations and deaths; demonstrate a significant decrease in antibody neutralization; reduce treatment effectiveness; and fail diagnostic detection (4, 5). In some cases, new variants emerge and then vanish, while in others, they emerge and then persist.
Etymologically, The COVID-19 Diary panelists explained that the World Health Organization (WHO) has a strategy for naming viruses that differs from previous practices of a virus being named after its country of origin or the scientist who first discovered it. Scientists now name viruses based on their structure, genome type (RNA or DNA), virus size, and whether it has a membrane, among other factors (2). The SARS-CoV-2 variants were thus named using the same criteria to avoid stigma and discrimination and to simplify public communications. The WHO convened a group of scientists from the Technical Advisory Group on Virus Evolution, the WHO COVID-19 reference laboratory network, representatives from A global initiative on sharing avian flu data (GISAID), Nextstrain, Pango, and additional experts in virological, microbial nomenclature, and communication from several countries and agencies to assign labels as names for SARS-CoV-2 variants, using Greek letters such as Alpha, Beta, Gamma, Delta among others (4, 6). Rambautet al. (year) also proposed a lineage naming system for SARS-CoV-2, which is now widely used; examples include B.1.1.7, B.1.351, and P.1 (7)
Alpha, also known as B.1.1.7, was the first of the highly publicized variants and the first VOC identified in the United Kingdom in December 2020 due to its higher transmissibility and pathogenicity (8,9). This variant became the dominant variant, and it is believed to be 30 to 50% more contagious than the original SARS-CoV-2. The B.1.1.7 lineage, according to scientists, is more likely to send infected people to the hospital and is deadlier than the original virus. Even though it is lethal, researchers believe that current SARS-CoV-2 vaccines can protect against the Alpha variant.
The Beta variant, or B.1.351, was discovered in South Africa at the end of 2020 and spread to other countries later. Experts were concerned about its multiple mutations and ability to evade antibodies. The beta coronavirus variant is approximately 50% more contagious than the original coronavirus. Though it does seem to spread more quickly than the original virus, it does not appear to cause more severe illness (10).
Delta, also known as B.1.617.2, is the most heard variant. This variant was discovered in India in late 2020 and quickly spread throughout the world. According to research, Delta is 40-60% more transmissible than Alpha and nearly twice as transmissible as the original SAR-CoV-2. Headache, sore throat, runny nose, and fever are all possible symptoms. (11)
Omicron, or B.1.1.529, is the next in line of the variant of the SARS-CoV-2 that was designated as a VOC after being discovered in Botswana and South Africa. This means that the variant may be more transmissible, cause more severe disease, and be less likely to respond to vaccines or treatments. On November 24, 2021, experts in South Africa reported the discovery of this variant to the World Health Organization (WHO) from a COVID-19 vaccinated patient, implying the new variant's immune invasion, and requesting updated vaccines (3,12). According to a study conducted by Columbia University researchers, the omicron variants are markedly resistant to the current COVID-19 vaccines, antibody treatments, and booster shots. There is still a scarcity of essential data regarding the infection rate to analyze its transmissibility. (13)
In addition to the Alpha, Beta, Delta, and Omicron variants, there are other variants that have been identified but classified as variants under monitoring (VUM) and variant of interest (VOI), indicating that they may pose emerging risks to global public health. These are the Gamma, Epsilon, Eta, Iota, Kappa, Zeta, and Mu variants (6, 8).
Virus mutation is a continuous process that results in the introduction of multiple variants. Though the latest variant's infectivity, prevalence, and severity are still unknown, investigations are underway to learn everything there is to know about the SARS-CoV-2 variants in order to recommend effective ways to prevent any upcoming outbreak (13). Therefore, previous recommendations to combat the spread of the COVID-19 pandemic must be followed religiously, including wash our hands with soap under running water, maintain social distance, and avoiding touching the eyes, nose, and mouth.
Author: Isabella Tetteh Ahinakwa (Host and Producer of HealthAfrik, Association of African Universities)
BIBLIOGRAPHY SOURCES CITED
1. https://escholarship.org/content/qt5kz643np/qt5kz643np.pdf
2. https://youtu.be/ZYH0-7ijhNQ
3. https://internews.org/wp-content/uploads/2021/02/COVID_Guidance_New_variants-English_0.pdf
5. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8256910/
6. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
7. https://media.nature.com/original/magazine-assets/d41586-021-00097-w/d41586-021-00097-w.pdf
8. https://www.yalemedicine.org/news/covid-19-variants-of-concern-omicron
9. https://journals.asm.org/doi/10.1128/Spectrum.01096-21
10. https://www.webmd.com/lung/coronavirus-strains
11. https://cdn.afresearchlab.com/wp-content/uploads/2020/03/02161836/Factsheet-Delta-Variant.pdf
13. https://onlinelibrary.wiley.com/doi/pdf/10.1002/jmv.27588




 SSNIT must be managed without gov’t interference – Austin Gamey
SSNIT must be managed without gov’t interference – Austin Gamey
 Ejisu by-election could go either way between NPP and independent candidate — Gl...
Ejisu by-election could go either way between NPP and independent candidate — Gl...
 We never asked ministers, DCEs to bring NPP apparatchiks for returning officer r...
We never asked ministers, DCEs to bring NPP apparatchiks for returning officer r...
 No one denigrated the commission when you appointed NDC sympathizers during your...
No one denigrated the commission when you appointed NDC sympathizers during your...
 Used cloth dealers protests over delayed Kumasi Central Market project
Used cloth dealers protests over delayed Kumasi Central Market project
 A/R: Kwadaso onion market traders refuse to relocate to new site
A/R: Kwadaso onion market traders refuse to relocate to new site
 Dumsor: Corn mill operators at Kaneshie market face financial crisis
Dumsor: Corn mill operators at Kaneshie market face financial crisis
 Jamestown fishermen seek support over destruction of canoes by Tuesday's heavy d...
Jamestown fishermen seek support over destruction of canoes by Tuesday's heavy d...
 Election 2024: EC to commence voter registration exercise on May 7
Election 2024: EC to commence voter registration exercise on May 7
 Public schools rebranding: We’re switching to blue and white, we’re painting all...
Public schools rebranding: We’re switching to blue and white, we’re painting all...
